Contact : enrique.carrillo@imdea.org
Technical Contact : david.perez@imdea.org
FooDrugs Introduction Video
E-Learning
This course aims to introduce the FooDrugs user in the importance of food - drug interactions and the usage of FooDrugs web tool to retrive infromation from the different tools
https://www.fns-cloud.eu/foodrugs/#/
FooDrugs help
1 Researchers
1.1-Food-CMAP
1.2-Gene sets
1.3-Food Transcriptomic studies
2 Clinicians
2.1 Text mining
Researchers
1-Food-CMAP
Video tutorial about how to explore information on similar gene signatures between foods and drugs
In this section you will have access to the potential FDI found by comparison of food transcriptomic
1.1 Query
Search for food/drugs and their interactions

There are three query options to choose from:
- Food-drug interactions Search for food/food compounds from GEO and get the interactions with CMAP compounds
- Food-gene interactions Search for food/food compounds from GEO and get the interactions with CMAP gene silencing/overexpression experiments
- Drug-food interactions Search for CMAP compounds and get the interactions with food/food compounds from GEO

Aditionally, for each query option you may specify which origin (cell line, tissue...) you want the query to be found in
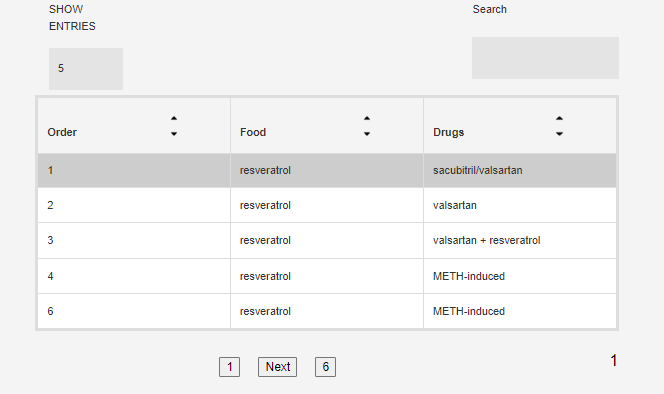
Each food/drug may have been tested in multiple conditions (origin, concentration, time point....) so below the search box you will see a table of all availabe conditions. Click on one to get the interactions for that particular condition

Tau threshold This parameter controls the strictness criteria for considering an interaction to occur. The higher it is, less but more confident interactions will be found
1.2 Enrichment analysis
Drugs and food compounds have been classified by numerous systems: drugs.com, drugbank, MeSH, cmap, kegg phytochemicals.
For each selected condition, run a binomial test on all classes for a given classification system to find out if a given food compound interacts positively/negatively more than expected by chance with a given class of drugs.
Run p value adjustement based on benjamini-hochberg or bonferroni
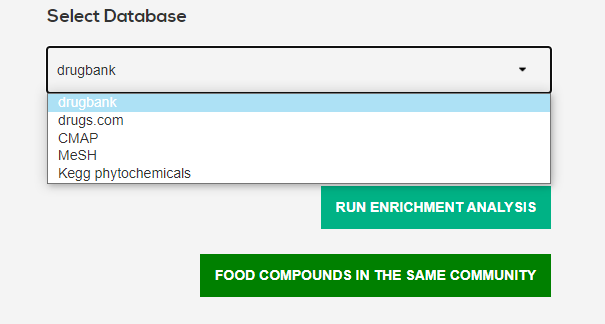
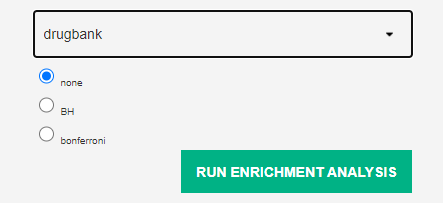
1.3 Community analysis
Network communities were found for interactions with a tau threshold of 90. Click on the button to access the communities of the selected condition

2-Gene sets
Video tutorial about how to explore information on similar gene signatures between foods and drugs
In this section you will have access to the top 150 UP and bottom 150 DOWN regulated genes obtained from differential expression analysis of food/food compounds in GEO. The differential expression was carried out with the typical limma + ebayes or voom + limma + ebayes methodology for microarray and RNA-seq data, respectively. These gene sets were sent to the CMAP to determone potential FDI
2.1 Query
Search for food/food compounds from GEO to get their gene sets
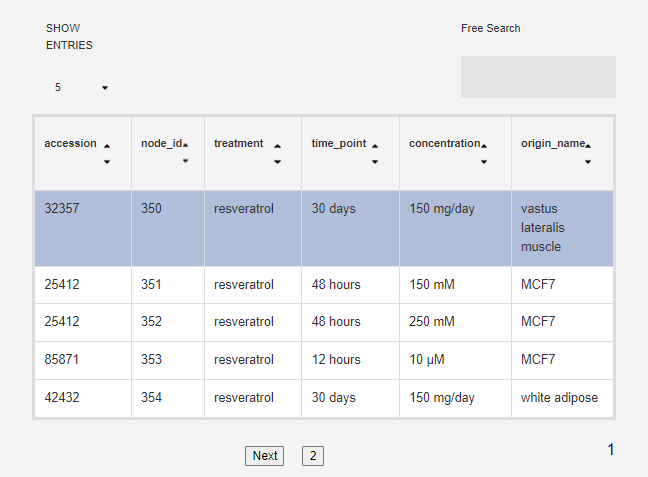
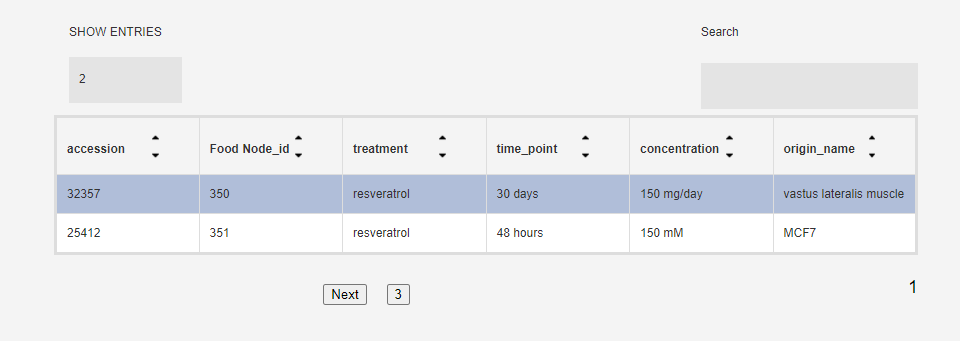
Each food/food compound may have been tested in multiple conditions (origin, concentration, time point...) so below the search box you will see a table of all availabe conditions. Click on one to get the gene sets for that particular condition
Aditionally, for each query option you may specify which origin (cell line, tissue...) you want the query to be found in
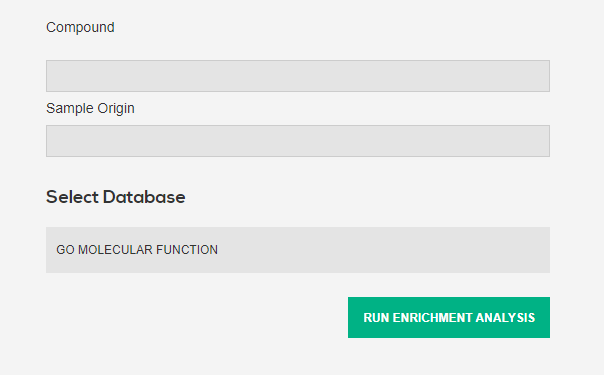
2.2 Enrichment analysis
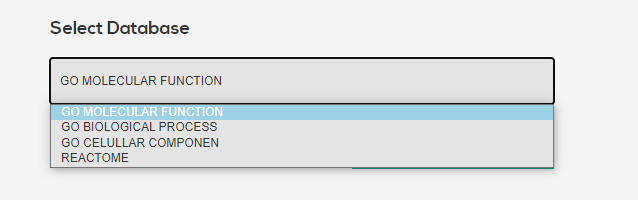
There exists many APIs for which to run enrichment analysis on gene sets. Available are all three gene ontology APIs, as well as reactome. More to come in the future. Click the button to send both UP and DOWN regulated gene sets to the respective APIs, with default options enabled.
3-Food Transcriptomic studies
In this section you will hace access to the GEO studies used for food/food compound trancriptomic analysis.
These studies were searched for using a keyword approach, collected from various food databases. Due to the high number of false positivies, unsupervised keyword extraction was done to manually search for promising candidates.
Next, metadata had to be manually extracted and normalized.
Metadata collected includes:
- Study metadata Title, platform, series number, study type, abstract
- Sample metadata Accession number, treatment, concentration, time point was extracted for all samples (not found is -). Additional metadata was also extracted for each specific study, not included in the results.
3.1 Query

Search for food/food compounds from GEO.

Each food/food compound may have been tested in multiple studies so the result will include all studies for that food/food compound.
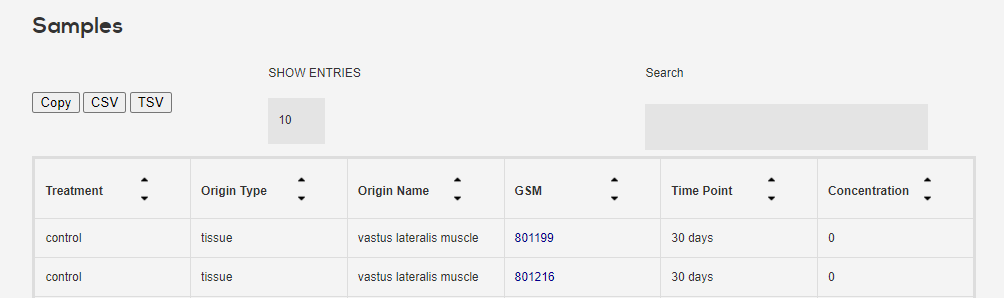
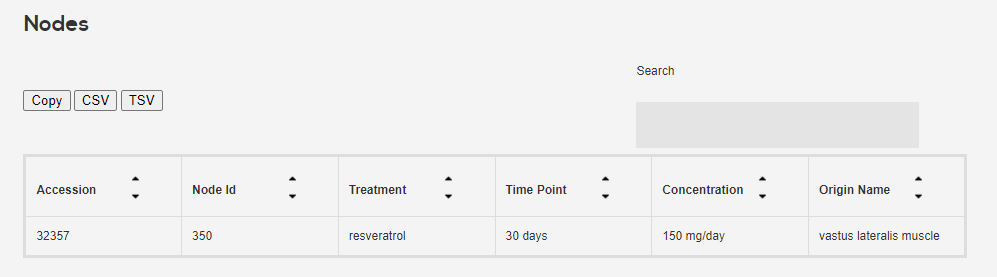
Click on one to get its sample information, as well as the nodes. Nodes are treatment-concentration-time point condtions which were analyzed for differential expression w.r.t to untreated control and sent to CMAP.

Aditionally, for each query option you may specify which origin (cell line, tissue...) you want the query to be found in.
Clinicians and nutritionists
1-Text mining
Video tutorial about how to explore potential food - drug interactions in text documents from the FooDrugs database
In this section you will have access to the FDI found by text mining on drugs.com, drugbank, medline and openFDA
1.1 Query
Search for any food, food compound or drug. Below the search box there will be all FDI found for that search term. Click on them to see the original text where it was extracted from, as well as a link to the source. The text mining system is not perfect, so there may be a few false positives